Computational and Mathematical Modeling Approaches to Intracellular and Intercellular Signal transduction
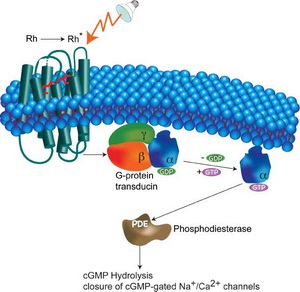
Signal transduction is the process of conversion of external signals, such as hormones, growth factors, and neurotransmitters, to a specific internal cellular response, such as gene expression, cell division, or even cell suicide. This process begins at the cell membrane where an external stimulus initiates a cascade of enzymatic reactions inside the cell that typically includes phosphorylation of proteins as mediators of downstream processes. Signal transduction consists of three processes. The first is reception – a ligand binds to a specific receptor on the cell membrane that triggers a change in the receptor molecule. The second is transduction. The change in the receptor brings about ordered sequences of biochemical reactions inside the cell that are carried out by enzymes and linked through second messengers. The third is response. After receiving the signal, target protein produces response which can be any of many different cellular activities, such as activation of a certain enzyme, rearrangement of the cytoskeleton, or changes in gene expression.
In our group, we use fundamental concepts from statistical physics and mathematical modeling to build an explicit understanding of both cellular and receptor scale signal transduction phenomena. The approaches of our works are to: building theoretical models of observed behavior based upon the physical laws that govern the atomic and molecular behavior of the receptors, examining the roles of receptor dimerization to the downstream signal, building mathematical models to describe the signal transduction pathway of G-protein coupled receptors, using Genetic Algorithm (GA) to optimize the signal transduction models to give the best fit with our own experimental results (in collaboration with Department of Physiology, Mahidol University) and studying the role of signaling noise to the cell decision. we use fundamental concepts from statistical physics and mathematical modeling to build an explicit understanding of both cellular and receptor scale signal transduction phenomena. The approaches of our works are to: building theoretical models of observed behavior based upon the physical laws that govern the atomic and molecular behavior of the receptors, examining the roles of receptor dimerization to the downstream signal, building mathematical models to describe the signal transduction pathway of G-protein coupled receptors, using Genetic Algorithm (GA) to optimize the signal transduction models to give the best fit with our own experimental results (in collaboration with Department of Physiology, Mahidol University) and studying the role of signaling noise to the cell decision.
Representative Research Publications:
-
Leelawattanachai J., Modchang C., Triampo W, Triampo D., Lenbury Y. Modeling and genetic algorithm optimization of early events in signal transduction via dynamics of G-protein-coupled receptors: internalization consideration. Appl. Math. Comput. 207(2) (2009), 528-544.
-
Modchang C, Triampo W*, and Lenbury Y. Mathematical Modeling and Application of Genetic Algorithm to Parameter Estimation in Signal Transduction: Trafficking and Promiscuous Coupling of G-protein coupled Receptors. Computers in Biology and Medicine, Vol. 38 (2008), 574-582.
-
Apiwat Wisitsorasak,Wannapong Triampo, Darapond Triampo, Charin Modchang, Yongwimon Lenbury. Stochastic biophysical model of heterodimerization of receptors: Monte Carlo simulations. (Submitted 2009)
-
Charin Modchang, Suhita Nadkarni, Wannapong Triampo, Herbert Levine, Wouter-Jan Rappel. A comparison of deterministic and stochastic simulations of neuronal vesicle release models.(Submitted 2009)
------------------------------------------------------------------------------------------------------------- |